Published on wheat.org.
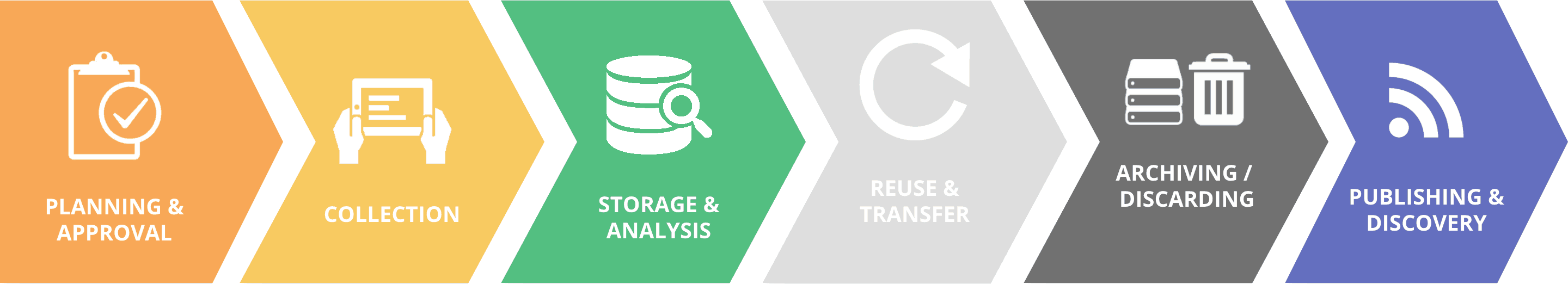
The MARPLE (Mobile And Real-time PLant disease) project – a project to test and pilot a revolutionary mobile lab in Ethiopia, led by the John Innes Centre, the International Maize and Wheat Improvement Center (CIMMYT) and the Ethiopian Institute of Agricultural Research (EIAR)—won the CGIAR Platform for Big Data in Agriculture Inspire Challenge Scale Up award in 2018.
We sat down with CIMMYT Principal Scientist and rust pathologist Dave Hodson to ask him about the project and its relationship with Big Data for Agriculture.
What is the significance of Big Data to your work?
Advances in sequencing technology, and the use of innovative big data approaches on sequence data from thousands of yellow rust isolates, opened the door for Diane Saunders and colleagues at the John Innes Centre in the UK to develop revolutionary, near-real time, mobile pathogen diagnostic techniques using portable palm-sized gene sequencers. The final result being the first operational system in the world using nanopore sequence technology for rapid diagnostics and surveillance of complex fungal pathogens in situ.
How do you see the role of the CGIAR Platform for Big Data in Agriculture in your work?
Support from the CGIAR Big Data Platform was critical to establish the partnership between John Innes, the Ethiopian Institute of Agricultural Research (EIAR) and CIMMYT and enable piloting and testing of the new MARPLE diagnostic platform in Ethiopia. The choice of Ethiopia to be the first country for initial testing was based on several key factors. Firstly, a strong national partner in EIAR; secondly, the critical importance of wheat and wheat rust diseases in the country. Ethiopia is the largest wheat producer in sub-Saharan Africa, but it is also considered the gateway for new wheat rust strains entering into Africa from Asia. All these factors made Ethiopia the highest priority country to take the lead in testing this revolutionary new and rapid pathogen diagnostics platform.
How did it impact this MARPLE project?
The pilot and subsequent scale-up project from the CGIAR Big Data Platform has enabled in-country capacity to be developed, and cutting edge technology for rapid pathogen diagnostics to be deployed in the front-line in the battle against devastating wheat rust diseases. The scientific innovation in developing the MARPLE platform, coupled to the suitability of the technology for developing country partners has now attracted support and interest from other donors. Matching funds were recently obtained for the scale -up phase of MARPLE from the Delivering Genetic Gain in Wheat project (implemented by Cornell University and funded by the Bill and Melinda Gates Foundation and the UK Department for International Development). This scale-up phase of the project will see a set of distributed MARPLE hubs established and embedded within the Ethiopian wheat research system – resulting in a sentinel system for the rapid detection of new yellow rust races that is unparalleled anywhere in the world. The scientific breakthrough in developing rapid diagnostics for complex fungal pathogens using nanopore sequencing will permit the development of similar systems for other important fungal diseases in the future.
Read full article on wheat.org →