Lab-in-a-backpack: Rapid Genomic Detection to revolutionize control of disease outbreaks in fish farming
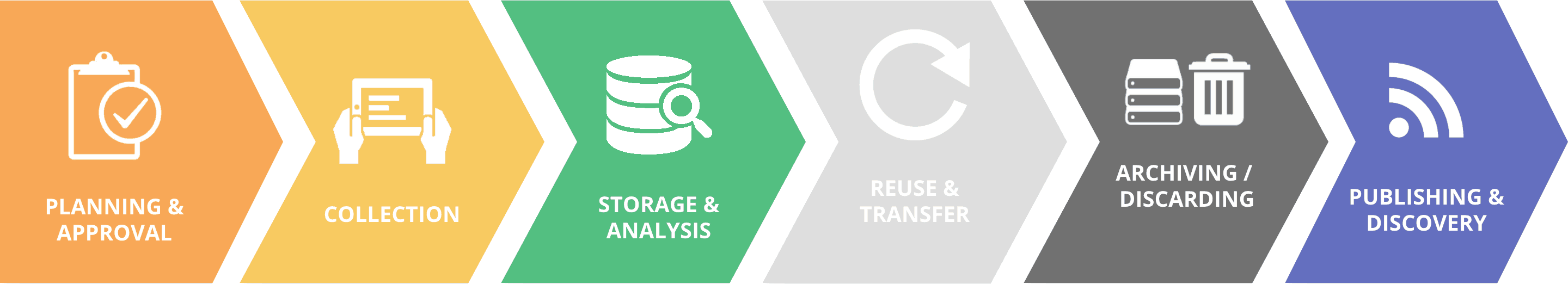
A winning 2019 Inspire Challenge project led by WorldFish, the University of Queensland, and Wilderlab is revolutionizing aquaculture disease control using big data approaches and genomic sequencing technologies.
A team of researchers from Malaysia, Australia, New Zealand, Thailand, and the UK is harnessing the power of genome sequencing to help Aquaculture farmers manage fish diseases. Until recently, this was not possible as the technology was not only costly but the genome sequencing process required time and coordination between different specialist labs. But this changed with the invention of MinION from Oxford Nanopore Technologies, a small and easy to use USB device that makes the genomic sequencing cheaper and faster.
The Rapid Genomics Sequencing team is now developing a cloud-based database to get high-resolution information from raw nanopore sequences allowing them to track outbreaks in close to real-time. The information they generate will then be accessed in a user-friendly format via a smartphone, enabling aquaculture inspectors to provide biosecurity advice and management plans to farmers in the palm of their hands. It will also inform local manufacturers what to put into custom vaccines and which farms need it. With real-time data accurately identifying what is killing the fish, local universities and start-ups will be able to produce simple vaccines for aquaculture health practitioners to vaccinate new stocks of fish before dissemination onto farms.

A field team prepares to collect samples from fish for disease diagnostic investigation.
In a demonstration at the WorldFish office, the team created a mock-up scenario of how this portable Lab-in-a-Backpack system works. If a fish farmer experiences a disease outbreak on his farm, local aquaculture inspectors collect samples from diseased fish and send them to a nearby lab where trained operators will be able to process them for whole-genome sequencing, with identification and typing of a bacterial pathogen taking just a few hours.
Click here to read the full article.
See the latest updates from the “Rapid genomic detection of aquaculture pathogens” project here.
March 30, 2020
CGIAR Research Program on Fish
Latest news