COPO: A portal to describe, store, and retrieve plant data
COPO, an open source tool to help researchers meet the FAIR data principles, aims to push the envelope on state of the art scientific data dissemination while minimising the burden of data publication and sharing.
Many recent efforts across the research data management spectrum have aligned with the FAIR principles; research data should be Findable, Accessible, Interoperable, and Reusable. Whilst much discussion is centered around what these principles mean and why they should be applied, there are relatively very few technical platforms trying to bridge the gap between what researchers need to do to meet their FAIR responsibilities and how they need to do it.
Implementing the FAIR principles in concrete systems approaches which can take into account the myriad of ways of recording metadata for the life sciences are lacking. Hence, we are developing COPO, an open source tool to help researchers meet these FAIR principles with a real world application.
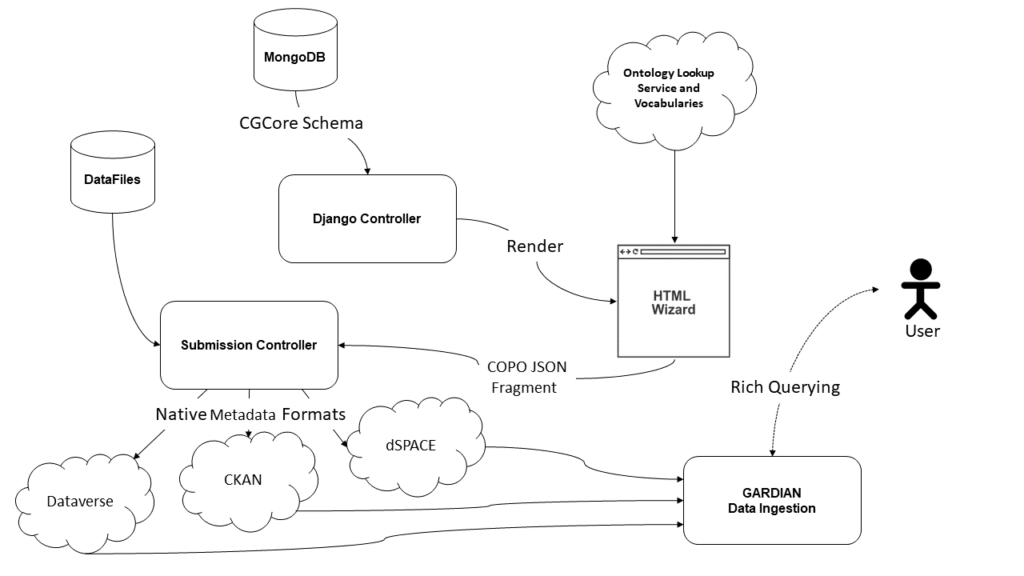
COPO is a web-based data brokering system that enables scientists to describe their research objects (raw or processed data, publications, samples, images, etc.) using community-sanctioned metadata sets and vocabularies, and then use public or institutional repositories to share them with the wider scientific community. COPO encourages data generators to adhere to appropriate metadata standards when publishing research objects, using semantic terms to add meaning to them and specify relationships between them. This allows data consumers, be they people or machines, to find, aggregate, and analyse data which would otherwise be private or invisible, building upon existing standards to push the state of the art in scientific data dissemination whilst minimising the burden of data publication and sharing.
COPO supports a range of datatypes and repository endpoints. Multi-omics data can be deposited to specialist repositories such as the European Nucleotide Archive, whereas more generic types such as spreadsheets, images, and documents can be submitted to either Figshare or a variety of off-the-shelf solutions such as Dataverse, CKAN, and DSpace.
Digital repositories used by CGIAR Centers are hosted locally, in some cases, and centralised in others, depending on the repository platform and the Center. Metadata for information resources (typically datasets and publications) are entered by the different data curators and information specialists of each Center. The objective of the CG Core metadata schema is to harmonise descriptions of agricultural resources across the CGIAR Centers through a minimum metadata set to maximise discoverability and interoperability. COPO provides a single entry point to the different systems to guarantee that the metadata are captured in a unified way across the different Center repositories. COPO user interfaces are generated based on the CG Core specification, with the advantage of providing data validation checks while the metadata are entered by the end user. COPO captures the full metadata set of the CG Core, although some data repositories only accept a subset of it. By retaining this metadata alongside links to these repositories, COPO improves the provenance of these records despite the full CG Core metadata not being supported by institutional repositories themselves.
COPO also supports the semantic tagging of both tabular data and free text data (in the form of Excel, CSV, and PDF files) with ontology terms, supplied in an auto-complete manner using the Ontology Lookup Service hosted at EMBL-EBI. Metadata templates can also be created by compositing ontology terms into a single set of fields, which can then be downloaded in spreadsheet format for completion and later parsing.
COPO is entirely open source and hosted under an MIT license on the Cyverse UK Infrastructure at the Earlham Institute in Norwich, UK.
Click here to access COPO.
Click here to access the freely available code on Github.
For more information, see our recent paper in F1000.
June 8, 2020
Felix Shaw
Research Software Engineer
Earlham Institute
16 - 16Shares
Latest news
16 - 16Shares