Webinar summary – Semantic annotation of images in the FAIR data era
The Ontologies Community of Practice hosted a webinar series to debate, share, and advance our thinking on selected topics in the domain of ontologies. Here’s what we learned from the third webinar.
***
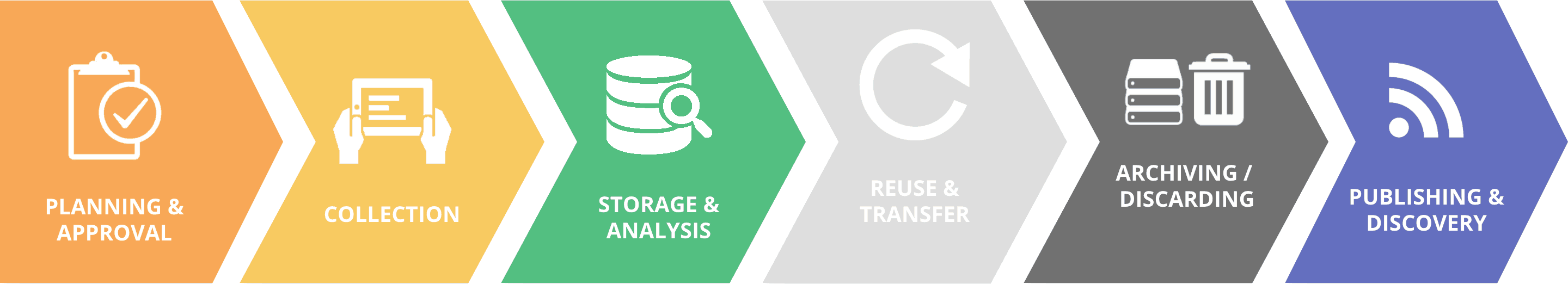
Digital agriculture increasingly relies on the generation of large quantity of images. These images are processed with machine learning techniques to speed up the identification of objects, their classification, visualization, and interpretation. However, images must comply with the FAIR principles to facilitate their access, reuse, and interoperability. As stated in recent paper authored by the Planteome team (Trigkakis et al, 2018), “Plant researchers could benefit greatly from a trained classification model that predicts image annotations with a high degree of accuracy.”
In this third Ontologies Community of Practice webinar, Justin Preece, Senior Faculty Research Assistant Oregon State University, presents the module developed by the Planteome project using the Bio-Image Semantic Query User Environment (BISQUE), an online image analysis and storage platform of Cyverse.
The Planteome module is a community platform for plant image segmentation and semantic classification using machine learning technics. The project focuses on leaf traits for which predicting detailed features with greater precision requires a large amount of data set training.
The segmentation of a photo is the identification of an area (e.g. a fruit) in the image that is then annotated using reference ontologies. Users can manually delineate an area of interest with markup lines, and then the module identifies it as a separated graphical object. Next, the trained model classifies the segment or the whole image to infer which object is represented. Ontology terms with their Universal Resource Identifier (URI) are then added into the metadata. The module currently uses a public image bank with 5,000 classified images.
By adding bootstrapping technics on a smaller set of training data, such a module could be used to identify visual disease symptoms. Around 300 images of leaves with fungal infection could be sufficient for tagging and classifying smaller sets of data. Semi-supervised technics can make multiplicative use of the data to improve the predictive power.
The next step of the Planteome project will be to improve knowledge representation with the integration of ontology graphs for better inferring image classification and improving interoperability. To this end, a metadata-querying interface is being developed. The current cutting-edge development is the real-time annotation of videos. Justin provides two examples of real-time annotation of maize kernel under fluorescent light and seed shape on the seed selector trail.
The Planteome module is on the Cyverse public repository, but it is not yet publically published. This is a collaborative project as the underlying system can take advantage of user-annotated images that contribute to the training set. Collaborators can propose their controlled vocabulary and contact the teams behind the reference ontologies for integrating their content.
Ontologies, which are powerful tools for knowledge representation and reasoning, can effectively support image tagging for indexing, retrieval, and content comparison. Pier Luigi Buttigieg, Data Scientist at the Alfred Wegener Institute, introduces the concept of “deep tagging” for a FAIRer future.
Knowledge representation is a branch of artificial intelligence that aims to model human knowledge into machine-understandable knowledge and support options for increased expressivity, advanced querying, and data mobilization. Knowledge representation for machine readability is key because enhancing tagging of information objects with a knowledge representation enables data discovery and, thus, new data analysis. Ontologies, when they are developed using the OBO Foundry principles, are the perfect tools for machine and human-readable logical representation of knowledge. Entities and relationships are defined on the web of knowledge along with their properties, processes, agents, etc. and possess a usable URI so as to be accessible to any human or machine. Ontology linked tags can add a new dimension of interoperability to metadata. High quality ontologies can also allow new kinds of analysis, driven by both data and machine-actionable knowledge. Community ontologies can be shaped by interacting with their developers.
The Planteome module follows as much as possible the FAIR principles; images are provided by public sources that are cited (e.g. Wikimedia) and described with quality metadata so they are findable, accessible, and reusable. Image annotation using community standard ontologies that are publicly available, going with the URI of terms, is a first step towards interoperability.
The future use of ontology graph will improve this criterion by offering the most expressive way of finding and reusing the images. One could then envisage a query system that can pull all relevant annotated images from various sources for answering a question such as, “Do we have images about what happens to the biodiversity surrounding a volcano after it has erupted?”
After a decade of ontology development, refinement, connections of concepts into semantic networks and data tagging, we are at a tipping point where the promise of knowledge inference and prediction is poised to become reality.
If you have a project for an image base and an analysis objective, please contact the Planteome team via their website.
Work cited: Trigkakis D. et al. (2018). Planteome & BisQue: Automating Image Annotation with Ontologies using Deep-Learning Networks. Proceedings of the 9th International Conference on Biological Ontology (ICBO 2018), Corvallis, Oregon, USA.
***
The Ontologies Community of Practice needs your expert knowledge to improve its content, so please let the team know of any comments, feedback, and suggestion for improvement. Contact information is available on the Ontologies homepage.
***
We thank the panelists for their engaging and inspirational insights:
Justin Preece
Senior Faculty Research Assistant at Oregon State University
Justin Preece designs and develops software for genomics research, including web and desktop applications, data transformation software, and databases. He currently works on the BISQUE project and the Planteome project that aims to provide reference ontologies for describing the function, traits, and phenotypes of plant genes, varieties, and germplasms.
Pier Luigi Buttigieg
Data Scientist at the Alfred Wegener Institute
Pier Luigi Buttigieg’s work focuses on the application of bioinformatics and multivariate statistics to the diverse data sets derived from microbial ecology investigations. Concurrently, he develops and co-leads the Environment Ontology (ENVO) and the Sustainable Development Goals Interface Ontology (SDGIO) in support of semantically consistent data standardization across the sciences and global development agenda.
***
The next webinar in this year’s Ontologies CoP webinar series will take place in December (exact date TBD). Please join to the Ontologies LinkedIn group and subscribe to the mailing list if you wish to receive more information and reminders on this topic.
***
Archived webinars can be found here.
September 10, 2019
Elizabeth Arnaud
Ontologies Community of Practice Lead
Bioversity International
Latest news