2019 Winner
Rapid genomic detection of aquaculture pathogens
Malaysia, Bangladesh
The project will pilot a readily deployable “lab-in-a-backpack” for pond-side identification and quantitation of pathogens affecting tilapia. Equipped with a portable DNA-extraction system, a hand-held DNA sequencer (MinION), a battery-operated minicomputer (MinIT), and an intuitive purpose-built software package, users without experience in molecular biology or bioinformatics will be able to identify fish pathogens from both water samples and infected tissues remotely and in real-time, with limited electricity and internet connectivity.
More about the project
Aquaculture is the world’s fastest growing food sector in the world and has been recognized for its potential to alleviate poverty and hunger. However, fish diseases and a lack of how to identify, track, and contain them can prohibit aquaculture development. This project will pilot a transportable, low-cost diagnostic “lab-in-a-backpack” that will enable users without molecular biology experience to identify fish pathogens in real-time with limited electricity and internet connectivity.
Aquaculture, the farming of aquatic organisms in both coastal and inland areas, accounts for 50 percent of the world’s fish that is used for food today. It is practiced by both some of the poorest farmers in developing countries and by multinational companies.
However, the development of aquaculture systems is often limited by fish diseases and a lack of knowledge and tools to identify fish pathogens, track their origin, and manage their spread.
Whole-genome sequencing informs how pathogens change and move through environments, permitting the implementation of evidence-based biosecurity to minimize disease impact.
Offsite sequencing services are expensive and cause prohibitive delays. Therefore, the project proposes leveraging offline supervised machine learning associated with the MinION portable sequencing device for low-cost diagnostics of fish pathogens in remote locations, allowing real-time disease investigation and data-driven management.
These tools will enable tilapia breeding, quarantine, and biosecurity centers, as well as academics and vets, to identify causal agents of disease outbreaks in a fraction of the time and cost required for external laboratory analysis; the project’s tests give results in hours rather than weeks or months and cost roughly 40 USD as opposed to more than 100 USD.
This Inspire Challenge proposal was selected as a 2019 pilot project winner, receiving a total of US$ 100,000 to put their ideas into practice. Learn more about the Inspire Challenge Grant here.
Step by step
2021
FEB 2021
Gates Foundation letter of intent
The team sent a letter of intent to the Bill & Melinda Gates foundation for the Global Grand Challenges on Smart Farming Innovations for Small-Scale Producers. The proposed project aims to create smart systems for antibiotic-free farm fish in Bangladesh in response to the challenge of production due to fish diseases directly impacting farmers’ livelihoods. If granted, the initiative will develop and implement transformational fish disease detection technology and prevention strategies for carp, tilapia and catfish, the most important crops for small-scale fish farmers.
FEB - JUNE 2021
Virtual training workshop
Using the samples collected from universities and private sector actors in Malaysia in October 2020, the team led a virtual training workshop on how to process, sequence, and upload genomic information to the cloud-based database.
MARCH 2021
Journal publication on fish diseases
The team published a paper detailing how Oxford Nanopore Technologies (ONT)-based amplicon sequencing is a promising platform to deploy in regional aquatic animal health diagnostic laboratories in low and medium-income countries, for fast and accurate confirmation and phylogenetic/genotyping of emerging infectious pathogens from field samples within a single day.
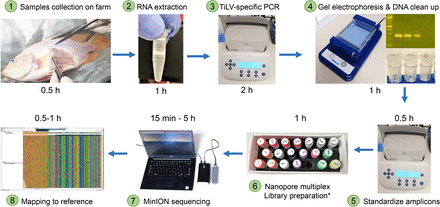
Overall workflow from sample collection of diseased fish on farm to sequence results. The entire process takes less than 12 hours. DNA repair, end-preparation, multiplex native barcode and adapter ligation.
APRIL 2021
Data analysis
The team is analyzing participants’ samples from past workshops and preparing all the content material for the virtual workshop that will be held in June 2021.
JUNE 2021
Virtual workshops
The team will hold workshops with academics, small-scale farmers from Bangkok, Bangladesh, and Malaysia on rapid diagnostics.
JUNE 2021
Developing a manual and training modules for field samplers
The team will build virtual training modules for field-based samplers composed of factsheets, short video tutorials, and easy-to-follow protocols for end-user software interfaces for the data outputs.
The manual will cover the entire process from biological sample collection to performing a sequencing run for analysis.
2020
JAN 2020
Bacterial genome sequencing and expansion of the team
The team sequenced 30 bacterial genomes and welcomed a new Ph.D. student, Suvra Das, from Bangladesh, to the team. Under Associate Professor Andrew Barnes at the University of Queensland, she will research processing methods for DNA extraction and library preparation to optimize the cost and performance of field sequencing tests.
Suvra Das, a PhD student in Andrew Barnes’ aquatic animal health laboratory, performs DNA extraction from fish pure bacterial isolates.
MARCH 2020
COVID-19 adaptations
Although the project was affected by the COVID-19 pandemic, the first year’s activities were primarily focused on laboratory and computer-based activities to generate the fish pathogen sequences, and, therefore, the team experienced fewer disruptions than field-based work.
However, various travel and workshop were adapted or postponed. A workshop that was originally planned to take place in person in Bangladesh has been converted to a virtual format and occurred in early 2021.
Watch the video below to hear from WorldFish Scientist Dr. Jerome Delamare-Deboutteville about the impacts of COVID-19 and progress throughout 2020:
JUNE 2020
Generation of aquatic pathogen genomic typing data
The team completed 50 bacterial genome sequences, generating two types of data:
- Highly accurate sequence data for all target aquatic pathogens derived from long and short-read sequencing was used to build the reference training database for machine learning algorithms.
- Raw nanopore read data for model development. This data was generated at the University of Queensland, Mahidol University/BIOTEC’s CENTEX Shrimp, and WorldFish.
Nurulhuda Ahmad Fatan from WorldFish demonstrates how to load a library onto the flow cell before starting a sequencing run on the Minion connected to the MinIT.
JULY 2020
Optimisation of field data acquisition and upload methodology
The team will compare sample collection and processing methods to optimise the cost and performance of the field sequencing workflows.
Sample extraction and library preparation and indexing methods will be compared to ensure that they can be completed in semi-remote locations.

Essential equipment needed to take single colonies from blood agar plates, extract DNA, and prepare a library for sequencing on the Minion, MinIT, and computer (or tablet or mobile phone) to visualise dashboard and launch a sequencing run.
JULY 2020
Building a software environment for typing pathogens from fuzzy data
To address the base-call error rate (<5 percent) of the MinION sequencing technology, the team developed a new bioinformatics software package that leverages machine learning to identify fish pathogens.
Two approaches were compared. In the first approach, hidden Markov models (HMMs) were used to compare experimental data to a reference database of hierarchical regions of differentiation. The second approach considered that all genomic regions provide information on strain type. Therefore, a rapid alignment method can be used to bin query samples probabilistically with the correct strain or type.
These models provided a position-specific scoring system that can account for base-calling inaccuracies and were trained on sequences from isoclinal pathogens obtained using the MinION.
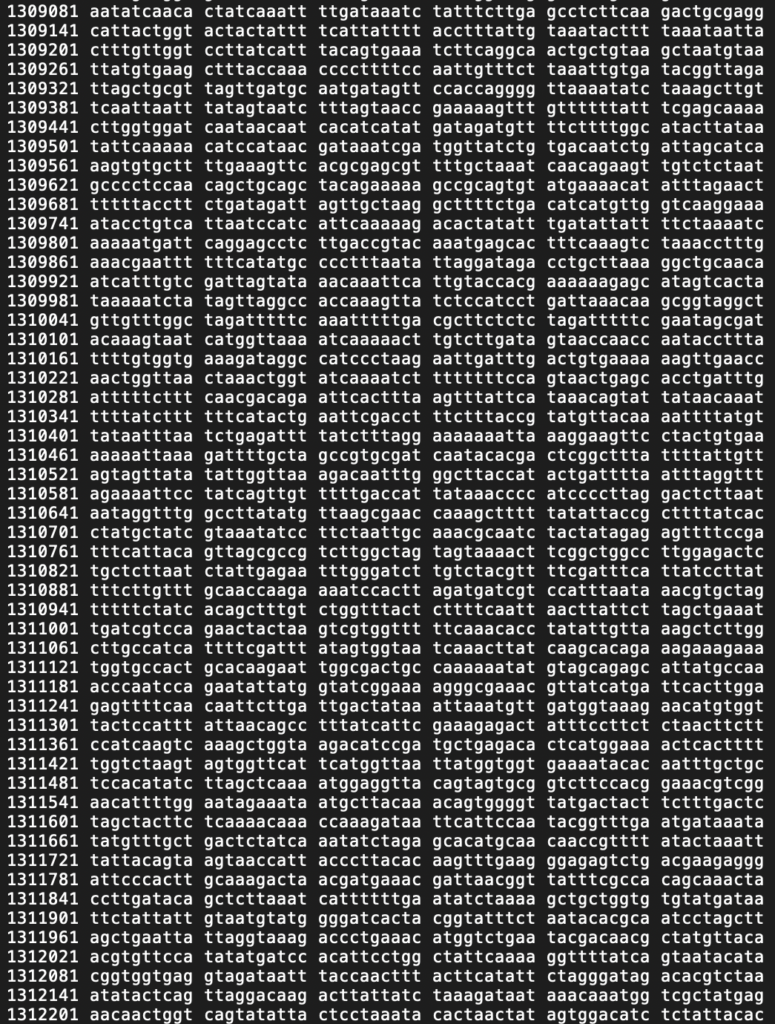
Example of an accurate genome sequence.
JULY 2020 - FEB 2021
Development of machine learning tools and cloud-based database
The team is creating a cloud-based database that features a large collection of fish pathogen genomes. The point-and-click user interface will be designed for public use, and the site is expected to be accessible in 2021.
OCT 2020
Sample collection
In place of in-person training workshops, the team collected samples from researchers at five leading universities and various private sector actors in Malaysia. These samples will be processed and used in an adapted virtual version of the workshop in early 2021.

A field team prepares to collect samples from fish for disease diagnostic investigation.
DEC 2020 - JUNE 2021
Engagement in joint initiative to increase aquaculture sustainability in Sub-Saharan Africa
Working through the WorldFish office in Egypt, the team is engaging 12 Master’s students (six from the College of Basic and Applied Sciences of the University of Ghana and six students from the College of Agriculture & Veterinary Sciences of the University of Nairobi) in a six-month intensive training on general aquaculture.
This effort is a part of a joint project led by WorldFish and the Norwegian Veterinary Institute to support aquatic animal health research, education, and management in Sub-Saharan Africa.
2019
OCT 2019
Project awarded US$100K Inspire Challenge grant
The project was one of four winners of the Inspire Challenge 2019 and was awarded US$100K at the Convention of the CGIAR Platform for Big Data in Agriculture, during 16-18 October, 2019.
Gender & Youth Inclusion
- WorldFish in partnership with GeneSEQ, UQ, and Wilderlab is working on the preparation of virtual training including sex-disaggregated data collected from the Malaysian participants involved in the project.